FWIW I think it would still be useful to get an issue opened about the BLAS/LAPACK stuff as it is more visible than being buried in a Discourse thread about various other topics. Sure, the path forward isn’t clear at the moment, but I think it is good to keep track of both the low hanging fruit and the bigger wishes in tandem, like we do for individual projects.
Yes, there are a lot of sub-optimal algorithms used. Part of the reason is that it requires specialized knowledge to know which algorithms to implement, as well as time to implement them.
For example, rectangular dilations and erosions can be computed in constant time wrt the kernel size. This is an algorithm known for 30 years or more. Yet not even OpenCV implements it.
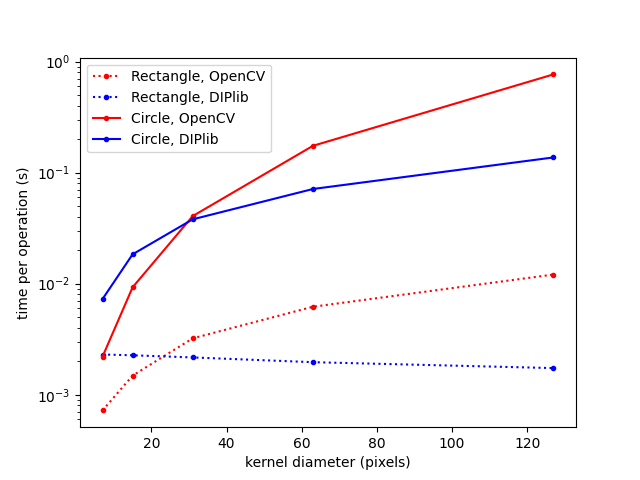
There are many similar examples. The median algorithm mentioned above is specifically 2D and uint8, it cannot be adapted to other cases. But there does exist another algorithm that is orders of magnitudes faster than the current skimage implementation and works for arbitrary dimensionalities and data types (ie more suited for skimage). I wrote about it here: Cris’ Image Analysis Blog | Median filtering
Implementing these algorithms for skimage or ndimage is hard though, I’ve already implemented them once in DIPlib, and have plenty left to do for that library, and little time to do it in. This is why you don’t see many PRs from me. ![]()
[By the way, I have tons of respect for the skimage maintainers. You guys are doing a great job! Don’t interpret any of this as derisive.]
I have a lot less experience with algorithms in other packages of this community, but am sure much of the same is true everywhere.
I find it interesting (and sometimes a bit worrying) that here as well as in many other discussions and contexts, SIMD is often mentioned without talking about cache, memory bandwidth, and maybe memory allocations at the same time. For data sizes exceeding cache sizes, many simple operations (addition, multiplication, matrix-vector multiplication, …) are actually bound by memory bandwidth and using SIMD has likely often limited effect on modern CPUs.
My nearly blind guess (with no overview of which operations are globally the most costly in scientific computing) would be that there is more to be gained (or at least more low-hanging fruit) by optimizing the use of caches and memory bandwidth instead of investing time in SIMD.
There’s benefit in both, it’s just that SIMD is a little easier to automate in some ways, given the potential for auto-vectorization, so there’s a bit more of an opportunity for cross-project infrastructure. It’s definitely not a panacea, as you pointed out, sometimes it ends up faster, sometimes slower, but being faster with no extra work is nice.
Optimizing for cache usage is more of a hands-on bespoke thing you need to do for each algorithm, and is definitely worth doing too.
I was sick for the past few weeks, but doing a bit better, and dropped a few more issues into Issues · scientific-python/faster-scientific-python-ideas · GitHub
If you have your own ideas, please add them there!
Expanded on the LAPACK topic mentioned above in Rewrite LAPACK so it's not in F77 · Issue #7 · scientific-python/faster-scientific-python-ideas · GitHub
I am late to this thread, but I have been working on optimizing up some Python packages like Pydantic and scientific packages like Albumentations and GDS Factory.
I have been building a new optimization tool called Codeflash that for pure Python code finds optimizations by replicating steps that an expert software engineer would follow.
The tool creates a performance benchmark or traces an existing one, generates correctness regression tests and line profiles to understand how a part of code behaves. Then the tool generates a variety of possible optimizations with LLMs and checks if they are indeed faster and correct. After it finds and proves a real optimization, it creates a pull request for you to review. Everything above is automated, and generates high quality optimizations. It works particularly well with optimizing algorithms, numerical code and logic.
If you would like to speed up a library, I would love to collaborate and volunteer to optimize it. I want to make Python ecosystem the fastest and would like to help as I can. Let me know how I can get involved.